Structural proteins
Whatever the complexity, size or shape of a virus particle, it is formed from an assembly of the structural proteins and serves to protect the viral genome from UV light or desiccation, which would otherwise fragment the nucleic acid, rendering the virus noninfectious. In naked virions, structural proteins form the capsid and (where present) the nucleocapsid. Enveloped virions also have structural proteins within the matrix and embedded in the membrane envelope. The structural proteins of a virion mediate virus delivery into cells and their major role is to provide attachment to cell receptors, as a first step to infection. Immune responses to these proteins often protect against viral infection and our understanding of this interaction forms the basis of modern vaccine design.
The icosahedral capsid of poliovirus is relatively simple with just four proteins, VP1, VP2, VP3, and VP4 (each cleaved from a polyprotein precursor), while the helical capsid of tobacco mosaic virus is simpler again, built with multiple copies of just one protein. In simple structures such as these, the capsid and nucleocapsid proteins can self-assemble to form the virion, but assembly of more complex capsids (such as that of herpes simplex virus or the double-shelled bluetongue virus) requires scaffolding proteins. These act as chaperones for the early association of the structural proteins, allowing the capsid to form, but they are then dismantled and play no part in the final virion structure.
Structural proteins must interact with the nucleic acid genome during the final stages of virion formation (assembly). Packaging of the genome (especially a large genome) into the confined space within a virion requires complex folding of the genome and intimate association through chemical bonding with one or more of the structural proteins. For example the N protein of rabies virus (Rhabdoviridae) and the NP protein of influenza virus (Orthomyxoviridae) both have positively charged amino acids that interact with the negatively charged RNA genome molecule, to encourage condensation (folding) of the RNA before packaging into the nucleocapsid. The viral proteins bind with secondary and tertiary structures in the genome that form as a result of specific nucleotide packaging sequences that anneal within themselves, folding the genome into regions of three-dimensional structure (not unlike the IRES structures of Class IV viruses). The correct association of structural proteins and packaging sequences becomes even more important when one considers the segmented genome of influenza virus, where eight different genomic RNA segments must each be packaged into a separate helical nucleocapsid, and all subsequently packaged within the final virion envelope.
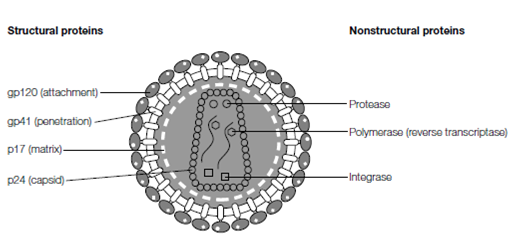
Figure: Virions are composed of structural proteins that enclose and protect the viral genome. The binding of structural proteins to cell receptors (attachment) initiates infection, delivering the genome into the target cell. Many viruses also carry nonstructural proteins in their virions; usually enzymes necessary for the first stages of virus replication. For example, HIV has two critical enzymes (reverse transcriptase and integrase) within its virion, and often a viral protease is present as well.
Virus envelopes contain structural glycoproteins, all of which must be targeted to the relevant cellular membrane before budding of the capsid. Many self-associate in small groups to form the tiny spikes (peplomers) visible on the envelope surface by electron microscopy, such as the hemagglutinin (HA, a trimer) and neuraminidase (NA, a tetramer) glycoproteins of influenza virus or the gp120 spike of HIV (a trimer; Table 1). The majority of a glycoprotein structure lies exposed on the outside of the envelope, being anchored within the envelope by a transmembrane domain. This part of the protein con- tains hydrophobic amino acids and laces back and forth across the membrane bilayer. Usually a relatively short region of the glycoprotein projects into the cell and virion interior, and is responsible for interaction with capsid or matrix (or tegument) proteins. The matrix proteins are not usually glycosylated but they may contain transmembrane anchor domains themselves. Hydrophobic regions can associate with the membrane or they may interact with the internal tail of the envelope glycoproteins.
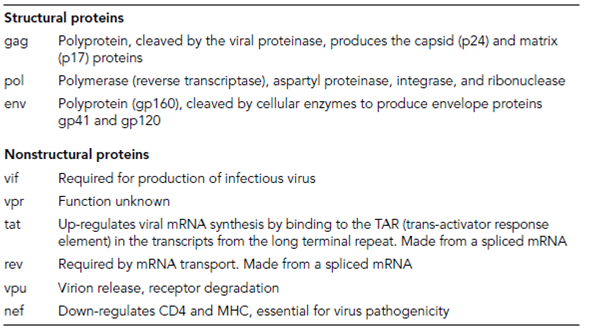
Table 1. Proteins of the human immunodeficiency virus (HIV) type 1