Types of viral genome
The viral genome carries the nucleic acid sequences responsible for the genetic code of the virus and, logically, larger genomes carry more genes, allowing these viruses to encode greater numbers of proteins, which may be structural (part of the virion) or non- structural. In infected cells the genome must be transcribed and translated into proteins, which requires the recognition of protein-encoding RNA molecules (messenger RNAs or mRNA) by cellular ribosomes. The processes of gene expression need to be appreciated in order to understand virus replication; they are well described in Section F of this book, and also more fully within the Instant Notes Molecular Biology volume. The virus genome, or a complementary copy of it, must also serve as a template from which new genomes can be synthesized; these ultimately assemble with viral proteins into progeny virions. Viral genomes can be of DNA or RNA in either single- or double-stranded form, although some viruses (e.g. hepatitis B virus) have strands of unequal size, so their genomes are only partially double-stranded. Genomes can be linear or circular in structure and some viruses spread their information between separate nucleic acid molecules (i.e. they can be segmented) that must all be present for successful infection of a host.
Virus genomes vary enormously in size; the smallest virus infecting humans is hepatitis B virus, with a genome size of just 3300 nucleotides, whereas some herpesvirus genomes are over 500 000 nucleotides. The largest virus genome sequenced to date is that of the marine mimivirus, at over 1 181 000 nucleotides; larger than the genome of some bacteria. The gene sequences within a virus genome must ultimately be translated into virus proteins and this requires either direct recognition of the viral genome as a messenger RNA (mRNA) by cellular ribosomes, or the conversion of viral genetic information into new mRNA transcripts within the infected cell. There are to date seven known schemes by which viruses provide mRNAs for translating their genomes into proteins and these distinct strategies form the basis of the Baltimore classification of viruses mentioned in the previous section and outlined in more detail below.
Double-stranded (ds) genomes are generally more stable, which is reflected in the range of sizes found for viruses of each type. The largest virus genomes are of dsDNA (e.g. Pox viridae, Herpesviridae, and the mimivirus mentioned above) with single-stranded (ss) genomes found in smaller sizes, although the use of segmented genomes allows more information to be encoded in multiple smaller (and hence less fragile) genome segments. As indicated above, all segments must of course be present in a single virion for infectivity. Single-stranded genomes are further categorized as being positive (+) sense or negative (-) sense. This refers to the relationship of the genome sequence to that of the mRNA representing its coding capacity. Where the genome sequence is the same (excepting the presence of thymidine instead of uracil) as that of the encoded mRNA, this is known as gene-sense or positive-sense. Conversely, genome sequences that are complementary to the mRNA transcript are known as anti-sense or negative-sense.
This has significance in ssRNA virus genomes; many that are (+) sense can be directly recognized as mRNAs and translated by cellular ribosomes immediately they enter a suitable cell. These genomes are said to be infectious as they are capable of initiating virus replication even in the absence of any virus proteins. Viruses with an infectious (+)ssRNA genome include poliovirus (Picornaviridae), SARS (Coronaviridae), yellow fever virus, dengue virus and hepatitis C virus (all Flaviviridae), and rubella (Togaviridae). A notable exception is the human immunodeficiency virus (HIV; Retroviridae), which has a noninfectious (+)ssRNA genome, and all negative-sense RNA genomes are noninfectious. The genomes of these viruses must first be transcribed into mRNAs and this requires virus enzymes (cellular enzymes cannot transcribe from an RNA template), which must be brought into the cells within the virus capsid. Such viruses include the influenza (Orthomyxoviridae), measles.
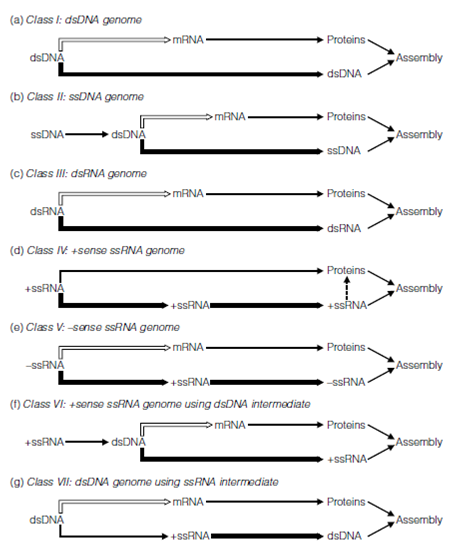
Figure: The Baltimore classification identifies several routes by which viruses with different genome compositions achieve mRNA transcription (shown as open arrows) and genome replication (thick arrows).
and mumps (Paramyxoviridae) viruses and the highly virulent rabies (Rhabdoviridae) and Ebola (Filoviridae) viruses. Some ssRNA viruses have regions of both positive- and negative-sense sequence and are known as ambisense genomes; they are noninfectious genomes, and include Lassa fever virus (Arenaviridae).