RNA viruses
All Class III virus genomes (dsRNA) are segmented. Transcription of mRNAs is from the genome molecules and requires a virally encoded RNA polymerase that must be packagedinto the virion for transport into infected cells. Only a few members of the Rotavirus and Orthoreovirus genera (Family Reoviridae) infect humans, but dsRNA viruses are a wide- reaching class and members have been found infecting animals, birds, fish, and insects and also plants, algae, fungi, and bacteria.
Class IV viruses all have positive (+) sense ssRNA genomes that can serve directly as mRNA transcripts for protein translation, and as such they possess a range of features that promote this role. Many (+)ssRNA genomes have 7-methylguanine, a modified ribonucleotide, bound at their 5¢ end. This molecule, known as the universal cap, protects the 5’ terminus of eukaryotic mRNAs and promotes mRNA translation by ribosomes in eukaryotic cells. In members of the Caliciviridae and Picornaviridae (e.g. poliovirus, hepatitis A virus) the cap molecule is absent, replaced by a small viral protein called VPg. Translation of these uncapped virus genomes requires the presence of additional sequences in the genome (between the 5’ end and the start of the coding region) that self-anneal to form a complex hairpin-like structure known as an internal ribosome entry site (IRES), which promotes translation of uncapped RNAs. Finally, the 3’ end of most Class IV genomes (again, like most eukaryotic mRNAs) is polyadenylated to ensure faithful protein synthesis.
During virus replication, negative-sense RNA copies of the genome are produced, from which mRNAs and new full-length genome molecules are transcribed. Transcription of all viral RNAs requires a virus-encoded RNA polymerase, synthesized rapidly in cells by translating the invading genome. Most Class IV genomes are small, ranging between 4000 and 13 000 nucleotides, although the SARS coronavirus (SARS-CoV ) and its relatives are much larger, reaching lengths of 28 000–31 000 nucleotides.Class V viruses all have (-)ssRNA genomes and many have segmented genomes. The genomes are not translated directly by ribosomes, so protein synthesis first requires transcription of mRNA from the genomic RNA; a few molecules of viral RNA polymerase are incorporated into the virion for this purpose. Interestingly, many viruses pathogenic to humans are found in this class, including the viruses responsible for mumps, measles, rabies, and influenza, and various hemorrhagic fevers such as Ebola, Marburg, and Lassa fever (the latter has an ambisense genome).
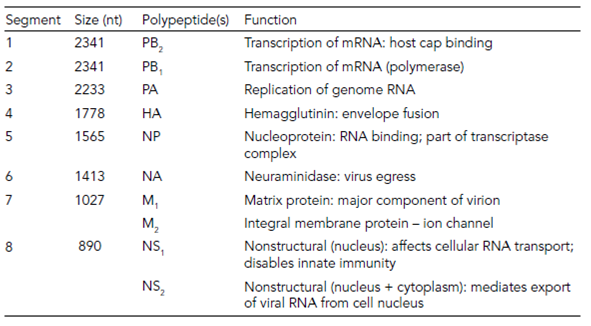
An important Class V family with segmented genomes is the Family Orthomyxoviridae, containing the influenza viruses, including those that infect humans. These viruses have seven or eight genome segments, each coding for one or two viral proteins (Table 1) and while a full complement of genome segments is necessary for infectivity, the source of each segment can be changed. This is known as reassortment, and allows the genomic segments of two different (but related) influenza viruses to swap, giving rise to new combinations. This sudden emergence of a ‘new’ virus genotype leads to the epidemics and pandemics of influenza that we have seen many times during our history.
On first appearance, Class VI virus genomes are like those of some Class IV viruses; they are of (+)ssRNA and possess both a universal 5’ cap and a 3’ polyadenylated tail. There are two RNA molecules within each virion, but they are identical and both carry the full coding potential of the virus, hence the genome is referred to as diploid, and is considered to be nonsegmented.
The genomic RNA, however, is not translated by ribosomes on entry into host cells but instead undergoes a complex replication strategy whereby it is first copied into a dsDNA molecule and then permanently cloned into the host cell chromosome. These steps require several virus-encoded enzymes (including a novel DNA polymerase, reverse transcriptase (RT), and an integrase) that must be brought into the cell along with the genome. Hence like Class V viruses, a few copies of these
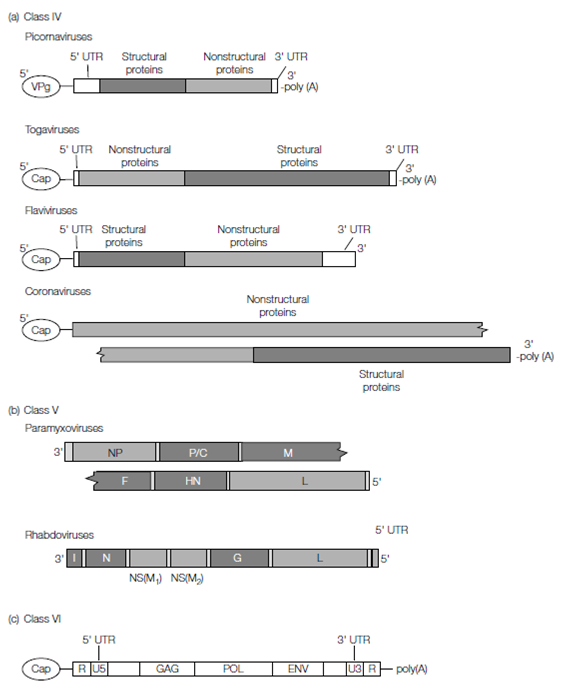
Figure: Diagrammatic representation of selected RNA virus genomes from (a) Class IV, (b) Class V, and (c) Class VI viruses. The position of structural and nonstructural genes is indicated, along with an indication of terminal structures or bound proteins. Untranslated regions (UTRs) do not encode viral proteins and usually contain regulatory sequences.
enzymes are found within the virion itself. Following integration of the dsDNA genome into a chromosome, only cellular enzymes are needed for transcription of viral mRNAs to allow viral protein synthesis to proceed. The most well studied virus family in the class is the Retroviridae, which includes the human immunodeficiency virus, HIV. More recent discoveries of related viruses in invertebrate and fungal species have been assigned to the families Metaviridae and Pseudoviridae.
Although discovered more recently, it is likely that these viruses are the ancestors of the vertebrate retroviruses.
The most recently described Class VII contains viruses reassigned from Class I and comprises only (to date) the Hepadnaviridae that involve hepatitis B virus, and the Caulimoviridae, which infect plants. These viruses have only partially dsDNA genomes (some regions are ssDNA) that are transcribed to mRNA by cellular RNA polymerase II once the genome has been made fully dsDNA. Like the Retroviridae they encode reverse transcriptase, which is active late during infection to synthesize new genomic DNA molecules from RNA templates. Hepatitis B virus has the smallest genome of any virus infecting humans (encoding only seven proteins from gene sequences) but it is estimated that there are 350 million people chronically infected with the virus. Clearly in the virus world, size (or complexity) isn’t everything.