DNA viruses
Class I viruses all have dsDNA genomes, which require the transcription of mRNA from the genome molecule; the virus may use the host cell RNA polymerase II enzyme (e.g. the Papillomaviridae, which can give rise to warts) or may encode its own (e.g. the Herpes¬ viridae). A few Class I viruses (those within the family Polydnaviridae) have a segmented genome but most have a nonsegmented genome.
Herpesviridae and Poxviridae are among the largest Class I viruses and both families contain members that infect humans. For example the herpes simplex virus genome is 152–154 kbp and that of cytomegalovirus is 235 kbp, while the now eradicated smallpox virus and its relative vaccinia virus have genomes of approximately 190 kbp. The large genomes of these viruses encode many proteins and their virions are correspondingly complex. The structure of herpesvirus genomes is unique in that several isomers of the same molecule can exist due to the presence of one or more regions of repetitive (identical) sequences, covalently joined to unique sequence regions. For example, within the genome of herpes simplex virus two unique sections (of differing length; a unique long (UL) and a unique short (US) region) are each flanked by inverted repeats, the terminal repeat long ( TRL) and terminal repeat short ( TRS) regions.
These repeats give the genome terminal redundancy and allow structural rearrangement of the unique regions that gives rise to four isomers, all of which are functionally equivalent.The genomes of Adenoviridae are smaller, with sizes ranging between 30 and 38 kbp and also have terminal redundancy; each strand has 100–140 bp of inverted repeat at the ends, which allows the denatured single DNA strands to self-anneal to form pan-handle structures, an essential step during replication. Adenovirus genomes are covalently
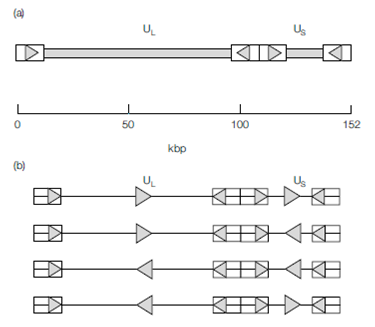
Figure: (a) Some herpesvirus genomes (e.g. herpes simplex virus) consist of two covalently joined sections of unique sequence, UL and US, each flanked by inverted repeats, represented by arrowed boxes. (b) This organization permits the formation of four different forms of the genome.
linked to a 55 kb terminal protein at the 5’ end of each DNA strand and this protein acts as a primer for the synthesis of new DNA strands.
The family Papillomaviridae (e.g. human papillomavirus 16 and 18, which are associated with cervical cancer) have very small genomes (about 8 kbp) that form a supercoiled circular structure and associate with four cellular histone proteins (H2A, H2B, H3, and H4). These viruses have between 8 and 10 genes, but even this small coding capacity is impressive for such a small DNA molecule and is achieved by several mechanisms. Firstly, papillomavirus genomes contain overlapping gene sequences and use shared promoter sequences (there are only seven within the genome). Secondly, the virus uses splicing to generate multiple, different mRNAs from individual primary RNA transcripts, a technique used by many viruses (RNA splicing was in fact discovered as a result of studies on virus genomes). Thirdly, these viruses encode proteins with multiple
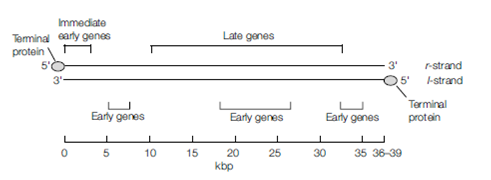
Figure . Organization of the adenovirus genome. A viral protein, the terminal protein, is covalently bound to one end of each of the DNA strands.
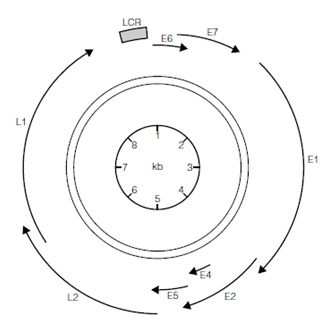
Figure: The organization of the small papillomavirus genome contains overlapping genes (arrowed lines) with shared promoters, including the regulatory sequences in the long control region, LCR.
functions, such as the E2 protein of human papillomavirus 16, which has functions linked with genome transcription, replication, segregation, and packaging. Many small virus genomes use these strategies and others, for example, Polyomaviridae use both DNA strands to encode proteins, so that a section of genome can be used in both directions. The larger virus genomes also use some or all of these strategies to an extent, but the majority of their genes are distinct and possess their own promoters.
Class II virus genomes are ssDNA and can be of positive or negative sense, and either circular or linear. All require the synthesis of a dsDNA intermediate from which transcription of their mRNA proceeds. Most ssDNA viruses are nonsegmented, but some are segmented. Furthermore, some viruses within the Begomovirus genus (Family Gemini¬ viridae) have two genomic segments that are packaged separately into two incomplete but joined icosahedral capsids.
Parvovirus genomes are linear nonsegmented ssDNA molecules of about 5 kb and like several of the Class I virus genomes they show terminal redundancy. The ends of parvo- virus genomes have palindromic sequences of about 115 nucleotides that self-anneal to form looped dsDNA regions, essential for the initiation of genome replication. Following genome replication, most newly assembled virions contain DNA strands of negative (-) sense, but some can contain positive (+) sense molecules. These very small genomes contain only two genes: rep that encodes proteins include in transcription and cap, that encodes the coat proteins. Originally only one parvovirus was known to infect humans (parvovirus B19), but since 2005 several new viruses have been identified, associated mostly with gastrointestinal infections.