AIS In Data Analysis's Application
The implementation of artificial immune systems in Data analysis model will consist of a set of cells, named antibodies, interconnected by associations along with linked connection strengths. The antibodies are assumed to shown the network internal images of the pathogens as input patterns contained in the environment to such this is exposed. The connections among the antibodies will find out their interrelations, giving a degree of similarity as in a described metric space in between them: the closer the antibodies, the more related as they are. The customized artificial immune network model can be formally explained as:
Definition
The Artificial immune systems based data analysis is an edge-weighted graph, not essentially fully liked, composed of a set of nodes, termed as antibodies, and sets of node pairs termed as edges along with an allocate number named as weight, or connection strength, connected with each connected edge.
The Artificial immune systems based data analysis clusters will serve as like internal images or mirrors causes for mapping existing clusters in the data set into network clusters. Because an illustration, assume there is a data set composed of three regions along with high density of data. The hypothetical network architecture will be produced by the learning algorithm.
The numbers inside the cells showing their labels or the sum of number is usually higher than the number of clusters and much smaller than the number of samples, and the numbers next to the connections represent their strengths, and dashed lines suggest connections to be pruned, in order to detect clusters and define the final network structure.
Notice the presence of three distinct clusters of antibodies, each of which with different number of antibodies, strengths and connections. These clusters map those of the original data set. Notice also that the number of antibodies in the network is much smaller than the number of data samples, differentiate the architecture proper for data compression. At last, the shape of the spatial antibodie's distribution follows the antigenic spatial distribution's shape.
There is no differentiating between the network surface molecules antibodies and their cells. The Ag - Ab and Ab - Ab interactions are quantified via proximity or similarity measures. The goal is to employ a distance metric to generate an antibody repertoire such constitutes the internal image of the antigens to be identified, and evaluate the similarity degree among the artificial immune systems based data analysis antibodies, such as the cardinality of repertoire could be controlled. Hence, the Ag - Ab affinity is inversely proportional to distance among them: the higher the affinity, the smaller the distance and vice-versa.
It is significant to stress such, in the biological immune system or IS, recognition happens throughout a complementary match among a specified antigen and the antibody. Nonetheless, in some artificial immune system applications, and for the intentions of this model, the creation of an antibody repertoire along with similar characteristics as instead of complementary to the antigen set is an appropriate alternative.
Because proposed in the original immune network theory or INT, the existing cells will contend for antigenic recognition and those successful will show the way to the network activation and cell proliferation, whereas those who fail will be removing. In addition, Ab - Ab recognition will conclude in network suppression. In our model, suppression is performed by removing the self-recognizing antibodies, specified a suppression threshold σs. Each pair Abj - Abi, j= 1, . . . M, i = 1, . . . , N, will concern to each other inside the shape-space S via the affinity dij of their interactions, that reflects the possibility of starting a clonal
The given notation will be adopted as:
- Ab : accessible antibody repertoire ( Ab ∈ S N × L , Ab = Ab{d } ∪ Ab{m} ) ;
- Ab{m} : whole memory antibody repertoire ( Ab ∈ S m × L , m ≤ N ) ;
- Ab{d} : d new antibodies to be placed in Ab ( Ab{d } ∈ S d × L ) ;
- Ag : antigen's population ( Ag ∈ S M × L ) ;
- fj : vector having the affinity of all the antibodies Abi (i = 1, . . , , N) along with relation to antigen Agj. So affinity is inversely proportional to the Ag - Ab distance;
- S: similarity matrix in between all pairs Abi - Abj, via elements sij (i, j = 1, . . . , N);
• C: population of Nc clones produced from Ab (C ∈ S Nc × L ) ;
• C*: population C after the affinity maturation process;
- dj : vector containing the affinity among every element from the set C*along with Agj;
- ξ : percentage of the mature antibodies to be chosen;
- Mj : memory clone for antigen Agj (stay from the process of clonal suppression);
- *Mj : for antigen Agj resultant clonal memory;
- σd : natural death threshold;
- σs : suppression threshold.
The artificial immune system or IS based data analysis learning algorithm intends at building a memory set that represents and recognizes the data structural organization. The more exact the antibodies, the less parsimonious the network or low compression rate, even as the more generalist the antibodies, the extra parsimonious the network with relative to the number of antibodies to enhanced compression rate. The suppression threshold (σs) controls or manages the antibody's clustering accuracy, specificity level and the network plasticity. The Artificial immune systems based data analysis learning algorithm can be illustrate in Programme no.2.
1. At iteration, do:
1.1 For each antigenic pattern
Ag j , j = 1, ... , M , ( Ag j ∈ Ag )
1.1.1 Find out its affinity fij, i = 1,...N, to all Abi - fu =1/Dij
Dij = ¦¦ Abi - Abj ¦¦,i = 1,...............N
1.1.2 A subset Ab{n} composed of the n highest affinity antibodies is chosen;
1.1.3 The n chosen antibodies are going to proliferate or clone proportionally to their antigenic affinity fi,j, producing a set C of clones : the higher or upper affinity, the larger the clone size for all of the n chosen antibodies.
1.1.4 The set C is submitted to a directed affinity maturation procedure or guided mutation producing a mutated set C*, where all antibody k from C* will suffer a mutation along with a rate αks inversely proportional to the antigenic affinity fi,j of its parent antibody : the higher affinity, the smaller mutation rate:
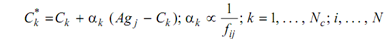
1.1.5 find out the affinity dkj= 1/Dkj among Agj and each elements of
C* : Dkj || C* - Agj ||, k = 1, . . . , Nc .
1.1.6 From C*, re-select ζ% of the antibodies along with highest dkj and put them keen on a matrix Mj of clonal memory.
1.1.7 Apoptosis: remove all the memory clones from Mj whose affinity Dkj > σd.
1.1.8 Find out the affinity Sik among the memory clones as:
Sik = || Mj,i - Mj,k ||, ∀ i, k .
1.1.9 Clonal suppression: remove those memory clones that Sik < σs.
1.1.10 Concatenate the whole antibody memory matrix along with the resultant clonal memory
Mj* for Ab{m} ←[Ab{m}; Mj*].
1.2 finds out the affinity in between all the memory antibodies from
Ab{m} : Sik = || Ab{m} - Ab{m} ||, ∀ i, k .
1.3 Network suppression: remove all the antibodies such that Sik < σs.
1.4 Build the whole antibody matrix
Ab{m} ← [ Ab(m) ; Ab{d } ] .
Programme no.2: Artificial immune systems Based Data Analysis Learning Algorithm