Initiation:
The whole mechanism of protein synthesis in eukaryotes is generally the similar as in prokaryotes, with three phases described as termination, elongation and initiation. Moreover, there are some significant differences, mainly in during initiation.
- While a prokaryotic ribosome has a sedimentation coefficient of 70S and subunits of 50S and 30S, a eukaryotic ribosome has a sedimentation coefficient of 80S with subunits of 60S and 40S. The composition of eukaryotic ribosomal subunits is also more complex than prokaryotic subunits but the function of every subunit is essentially the same as in prokaryotes.
- In eukaryotes, each mRNA is monocistronic which is, discounting any subsequent post-translational cleavage reactions which should occur the mRNA encodes a one protein. In prokaryotes, several mRNAs are polycistronic which is they encode many proteins. Each coding sequence in a prokaryotic mRNA has its own termination and initiation codons.
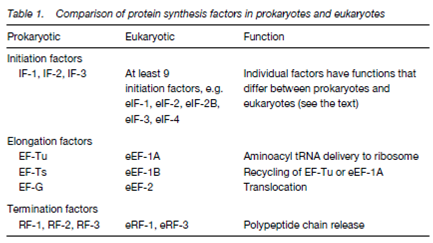
- Beginning of the protein synthesis in eukaryotes needs at least nine distinct eukaryotic initiation factors (eIFs) compared with the three initiation factors (IFs) in prokaryotes.
- In the eukaryotes, initiating amino acid is methionine isnot N-formylmethionine as in prokaryotes.
- As in prokaryotes, a special initiator tRNA is needs for initiation and is distinct from the tRNA which recognizes and binds to codons for methionine at internal positions in the mRNA. When charged with methionine prepared to start initiation, this is known as Met-tRNAimet.
- The major difference among initiation of translation in eukaryotes and prokaryotes is that in bacteria, a Shine-Dalgarno sequence lies 5' to the AUG initiation codon and is the binding site for the 30S ribosomal subunit marking this AUG as the one to use for initiation rather than any other AUG internal in the mRNA. The starting complex is assembled straightly over this initiation codon. In compare, most eukaryotic mRNAs do not hold Shine-Dalgarno sequences. Alternatively, a 40S ribosomal subunit attaches at the 5' end of the mRNA and moves downstream instance for in a 5' to 3' direction until it finds the AUG initiation codon. This procedure is known as scanning.
The overall details of initiation in eukaryotes are still not fully known but the process occurs widely as follows:
- The first step is the formation of a pre-initiation difficult consisting of the 40S small ribosomal subunit, Met-tRNAimet, GTP and eIF-2;
- The pre-initiation complex now binds to the 5' end of the eukaryotic mRNA, a step that needs eIF-4F which is also called eIF-3 and cap binding complex. The eIF-4F complex consists of eIF-4E, eIF-4A and eIF-4G; eIF-4E binds to the 5'cap on the mRNA (an essential step) whilst eIF-4G interacts with the poly (A) binding protein on the poly (A) tail (that of course implies in which the mRNA bends back on itself to permit this interaction to occur). Therefore this complex checks which both the 5' and 3' ends of the mRNA are intact. An eIF-4A is an ATP-dependent RNA helicase which unwinds any secondary structures in the mRNA, ready it for translation. This is in contrast to prokaryotic translation where no helicase is required, presumably since protein synthesis in bacteria can begin even as the mRNA is since being synthesized while in eukaryotes transcription in the translation and nucleus in the cytoplasm are separate events that permits time for mRNA secondary structure to form.
- The complex now moves along the mRNA in a 5' to 3' direction until it locates the AUG initiation codon like scanning. The 5' untranslated regions of eukaryotic mRNAs vary in length but can be many hundred nucleotides long and may contain secondary structures like as hairpin loops. Those secondary structures are probably erased through initiation factors of the scanning complex. The initiation codon is commonly recognizable since it is frequent (but not always) contained in a short sequence is known the Kozak consensus (5'-ACCAUGG-3').
- Once the difficulty is positioned over the initiation codon, the 60S huge ribosomal subunit binds to form an 80S initiation complex, a step which needs the hydrolysis of GTP and leads to the release of various initiation factors.