Promoters
RNA polymerase is composed of four different polypeptides called beta, beta prime, alpha and the sigma factor(αββ'σ). The core polymerase(αββ') has a high processivity but low DNA affinity. The addition of the sigma factor to make holopolymerase confers DNA sequence specificity, forcing the RNA polymerase to bind at the promoter region. Once the RNA polymerase has bound the sigma factor dissociates. The core polymerase then synthesizes RNA complementary to the lower DNA strand.
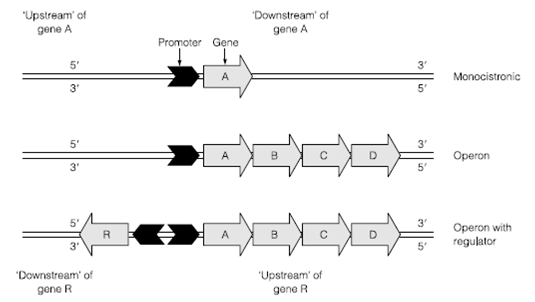
Figure: The arrangement of genes and promoters in Bacteria.
The promoter region tends to have a slightly higher A and T content than the surrounding DNA and the most abundant form of promoter in Escherichia coli has two fairly well conserved sequences TATAAT (the Pribnow box or –10 sequence) and TTGACA (the –35 sequence). The higher A or T content means that there are fewer hydrogen bonds to hold the double-stranded DNA together and thus the double helix is easier to force apart a process termed melting. Melting of the promoter region allows access of the RNA polymerase which specifically targets the –10 and –35 regions through the use of the sigma factor. The numbers –10 and –35 refer to relative position in relation to the number of bases the sequence is from the base where transcription starts known as the transcription start site.
The –10 (TATAAT) /–35 (TTGACA) consensus promoters is not the only sequence of promoter in the genome. There are several promoters with a sequence very similar to –10/–35 and the greater the difference in the base sequence is to this consensus the weaker the promoter. A strong promoter like as that of the lac operon forms a very tight bond with the sigma factor and transcription is very likely to be initiated from like a promoter. A weak promoter binds the sigma factor through only a few bases and is concomitantly less likely to initiate RNA synthesis. Other promoters have completely different sequences that bind alternative sigma factors. A good example of this is the alternative sigma factor produced in response to low oxygen in E. coli and some other bacteria. The sigma factor still allows RNA polymerase to recognize a –10 site but instead will only allow the holoolymerase to bind where there is an additional specific sequence at around 42 bases upstream of the transcription start site. The use of alternative sigma factors allows a whole group of genes and operons known as a regulon to be switched on and off according to external changes in the cell’s environment. Induction of the FNR regulon allows the cell to induce all the genes that are useful to cope with low oxygen, principally alternative electron acceptors.