Allosteric enzymes
Against [S] a plot of V0 for an allosteric enzyme provides a sigmoidal curve rather than the hyperbolic plots predicted through the Michaelis–Menten equation. The curve has a steep section in the center of the substrate concentration range refiecting the rapid increase in enzyme velocity that occurs over a narrow range of substrate concentrations. This permits allosteric enzymes to be particularly sensitive to little changes in substrate concentration within the physiological range. In allosteric enzymes binding of the substrate molecule to one active site affects the binding of substrate molecules to other active sites in the enzyme; the variant active sites are said to behave cooperatively in acting and binding on substrate molecules cf. binding of O2 of hemoglobin to the four subunits. Therefore allosteric enzymes are frequent multisubunit proteins with one or more active sites on every subunit. The binding of substrate at one active site induces a conformational modification in the protein which is conveyed to the other active sites altering their affinity for substrate molecules.
Two models have been put forward to account for the allosteric effects observed in proteins. In the concerted or symmetry model, first described through Jeffries Wyman, Jacques Monod and Jean-Pierre Changeaux who sometimes known to as the Monod-Wyman-Changeaux (MWC) model the subunits of an allosteric enzyme can exist in one of only two states that are T and R. T-state subunits are in a tense state which is relatively and compact inactive, therefore R-state subunits are in a expanded, relaxed, active state with higher affinity for the substrate; no intermediate states are permitted. In the absence of bound substrate the equilibrium favors the T-state. As substrate binds to every active site in the T- state the equilibrium shifts towards the R-state. All of the subunits modify conformation in a concerted manner, that implies in which the conformation of every subunit is constrained through its association with the other subunits; in other
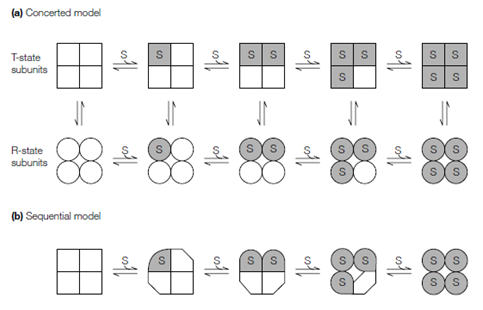
Figure: Models of allosterism. (a) The concerted or symmetry model; the squares and circles represent the T- and R-states, respectively. (b) The sequential model; substrate binding progressively induces conformational changes in the subunits.
words, there are no oligomers which concurrently contain R- and T-state subunits and the molecular symmetry of the protein is conserved in during the conformational modification.
In the alternative sequential model which is first proposed through Daniel Koshland, sequential modification in structure take place within an oligomeric enzyme as the individual active sites are engaged. The binding of substrate to one site infiuences the substrate affinity of neighboring active sites without necessarily inducing a transition encompassing the overall enzyme, such that the molecular symmetry of the overall protein is not necessarily conserved. The sequential model builds upon of enzyme–substrate interaction the induced-fit hypothesis, although the concerted model implicitly assumes the lock-and-key model of substrate binding to the enzyme’s active site. In the sequential model, substrate binding induces a conformational modification in a cooperative and subunit interactions arise by the infiuence in which these conformational changes have on neighboring subunits. Strengths of these interactions depend on the degree of mechanical coupling among subunits. In the sequential model the enzyme–substrate binding affinity varies with the number of bound substrate molecules, although in the concerted model this affinity depends only on the quaternary state of the enzyme. The results of learning of a number of allosteric proteins suggest in which most behave according to a combination of the sequential and concerted models.
Allosteric enzymes may be controlled through effector molecules activators and inhibitors which bind to the enzyme at a site other than the active site either on the similar subunit or on a various subunit, thus causing a modification in the conformation of the active site that alters the rate of enzyme activity cf. the binding of H, CO2 and two, three-bisphosphoglycerate to hemoglobin.An allosteric activator raises the rate of enzyme activity, although an allosteric inhibitor reduces the activity of the enzyme.