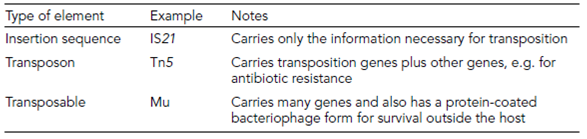
Transposition
Transposition is the insertion of short DNA fragments into any position in the genome, a process carried out by transposons, insertion sequences (IS), and some specialized phages ( Table 1). This is sometimes called illegitimate recombination as it appears to require no homology between sequences. However, analysis of transposon positions after transposition suggests that it is a pseudo-random event, and that the recombination is site-specific. However, the recognition sites are extremely degenerate and occur so frequently in most genomes that almost all genes can be interrupted by the entry of a transposon.
The presence of insertion sequences can be detected throughout the genomes of many Bacteria and Archaea as well as in their plasmids, and they are essential to some functions. The integration of the F? plasmid into the E. coli chromosome is mediated by homologous recombination between identical insertion sequences. The ecology and microbiology of transposons is extremely complex, given that some of these elements can move around the genome in the absence of chromosomal replication and initiate a form of
Table 1. Transposable elements in E. coli
conjugation to transfer themselves from one cell to another. In some senses, transposable elements could be looked on as very basic viruses especially given that transposition is the method of replication for bacteriophage such as Mu.
A simple transposition event is illustrated by the conservative transposition of an IS element. The IS encoded transposase makes cuts at another site in the host chromosome (in a pseudo-random manner) and is excised and integrated into the new site in much the same way as l achieves lysogeny. The IS is flanked by inverted terminal repeats, which aid in the transposition process. As the transposase cuts are staggered, the bases either side of the repeats are duplicated after integration.
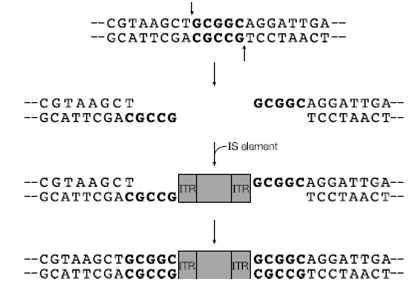
Figure: Transposition of an insertion sequence (IS) element into a host DNA with duplication of the target site (shown in bold)
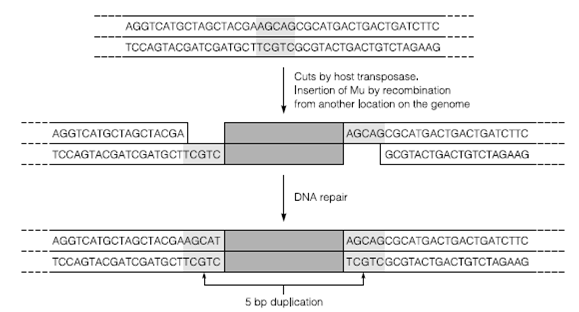
Figure 3. Insertion of bacteriophage Mu by transposition into a bacterial genome
The transposable viruses do not excise themselves during transposition, but place a copy elsewhere on the genome instead. This process of replicative transposition allows Mu to replicate its genome in the host in terms of copy number per chromosome as well as during cell division.
Transposons offer a convenient method of generating mutants, particularly when gene interruption is required. Transposon mutagenesis is most developed for Tn5 derivatives, in which additional genes such as those for kanamycin resistance or b-galactosidase activity have been added. A bacterial strain can be transformed with a plasmid bearing the modified transposons. The transposons can then jump from the plasmid to the chromosome and cause random interruption of genes. Auxotrophs or other mutants can then be identified as normal.