Homologous recombination
This process is sometimes called generalized recombination and involves the exchange of DNA between two regions of homology (i.e. similar but not necessarily identical DNA sequence) on two different DNA molecules. One of these regions might be on a plasmid, the other on the chromosome. In rare instances these two regions could be on the same chromosome but many hundreds of base pairs apart or more. After recognition and alignment of the regions of homology by the enzyme RecA a protein complex, RecBCD, digests dsDNA until it reaches a chi sequence (crossover hot insti- gator, sequence GCTGGTGG). The activity of RecBCD then changes to a single strand- specific 5'–3' exonuclease, so that a single DNA strand remains as a long 3' overhang. This is represented by the arrowheads in. The free single-stranded DNA becomes coated with RecA protein, and can migrate into the other DNA molecule in a process of strand invasion. Once the nicks in the DNA strands have been joined, the two
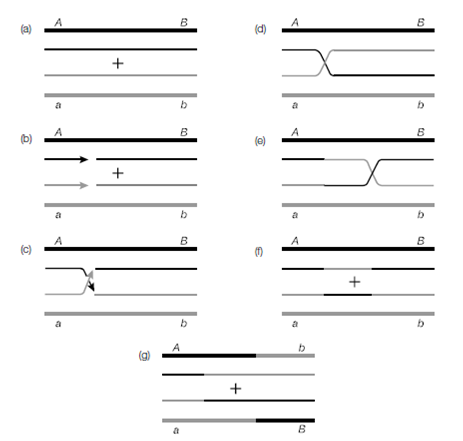
Figure: The Holliday model for homologous recombination.
DNA molecules adopt a cross-shaped formation known as the Holliday junction and is named after the person to first propose it.
The binding of RuvA to the Holliday junction leads to the binding of RuvB to adjacent double-stranded DNA. RuvB is a translocase, and promotes the movement of the Holliday junction along the DNA molecules. This is known as branch migration. The single-stranded parts of the junction are cleaved (strand exchange) and the two molecules become separate molecules again (resolve) when RuvC binds to the RuvAB–DNA complex. How the resolution proceeds is still the subject of some debate, but it has been postulated that equilibrium is established between a RuvA planar Holliday junction and tetrahedral junctions stabilized by another protein, RusA. Depending on the direction of a final cleavage, there are two possible outcomes.