Plasma membrane proteins:
Integral plasma membrane proteins are also synthesized through ribosomes on the RER, but become inserted in the rough endoplasmic reticulum membrane rather than transported into the lumen. In During transport to the Golgi and then to the cell surface these proteins stay anchored in the membrane, the last vesicles that fuse with the plasma membrane then becoming new plasma membrane that is shown in the below figure. Note that, after insertion
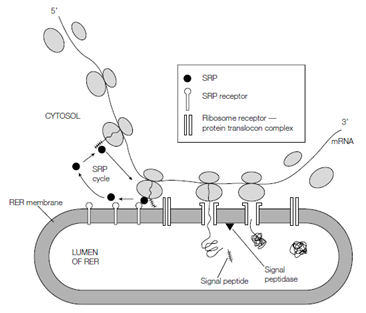
Figure: A simplified version of the signal hypothesis (see the text for details).
In the RER membrane, one part of the protein faces in towards the RER lumen but eventually this faces outward on the cell surface. It is this category of the protein that receives the carbohydrate during glycosylation in the RER and Golgi complex so that the carbohydrate is exposed on the cell surface.
The Transfer of the plasma membrane protein across the ER membrane happens during synthesis through a mechanism same to that for secretory proteins. Moreover, through definition, the protein is destined to remain anchored in the membrane and not enter the RER lumen entirely. There are various ways in that this is achieved, depending on the kind of membrane protein. Several integral membrane proteins are one membrane-spanning proteins, which is the polypeptide chain crosses the membrane only once, while in other cases the protein is a multiple membrane-spanning protein.
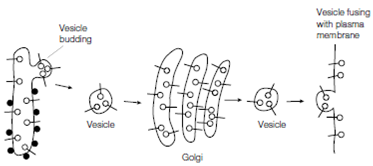
Figure: Synthesis of plasma membrane proteins; see text for details. The ribosomes attached to the RER are shown as filled-in circles whereas the newly synthesized plasma membrane proteins are shown as open circles on stalks.
The orientation of the protein in the membrane and the number of times it spans the lipid bilayer depend on specific topogenic series within the polypeptide chain. These topogenic series are regions of predominantly hydrophobic amino acids and fall into three categories: internal signal sequences, stop-transfer sequences and N-terminal signal sequences.
In the single membrane-spanning Type I integral membrane proteins shown in the figure, Additionally to the N-terminal signal sequence that is cleaved from the protein through signal peptidase as in secretory proteins, there is a 2nd hydrophobic sequence situated internally in the protein. Therefore the protein begins to cross the RER membrane during synthesis such as a secretory protein, but then transfer is end before the entire protein is translocated and the protein stays inserted in the membrane through the interaction of the hydrophobic stop-transfer sequence with the hydrophobic interior of the bilayer. In the one or single membrane-spanning Type II membrane proteins, there is just an N- terminal signal sequence as establish in secretory proteins. Moreover, in this case the signal sequence is not cleaved from the membrane protein through signal peptidase and doubles as the membrane anchor. The Multiple membrane-spanning Type III integral membrane proteins, that cross the membrane various times, have multiple internal signal peptides and stop-transfer sequences to organize this arrangement during synthesis. The last orientation of the N terminus and the C terminus depends on whether the N-terminal signal sequence is whether and cleaved the last topogenic sequence is an internal signal sequence or a end-transfer sequence, respectively. Some proteins lack an N-terminal signal sequence and have just an internal signal sequence.
Those proteins which are anchored in the membrane by a covalently attached GPI (glycosylhosphatidylinositol) structure at the C terminus possess both an N-terminal signal sequence to direct them to the RER membrane and a second hydophobic sequence at the very C terminus. The N-terminal signal sequence is cleaved off through signal peptidase, although the C-terminal sequence directs the addition of a preformed GPI structure to an internal amino acid residue near the C terminus. The GPI structure is built up through sequential addition of sugars (glucosamine and mannose) and ethanoloamine phosphate to phosphatidylinositol in the RER membrane. The transamidase enzyme then cleaves off the C-terminal signal sequence and concomitantly adds on the complete GPI anchor.