Sequencing strategy
An ‘average’ sized protein of 50 kDa would contain around 500 amino acids. Therefore, even with large amounts of highly purified material only about the N-terminal one-tenth of the protein can be sequenced through Edman degradation. In order to series a larger protein, the first phase is to cleave it into smaller fragments of 20–100 residues which are then sequenced and separated. Specific cleavage can be achieved through enzymatic or chemical techniques. For instance, the CNBr (chemical cyanogen bromide) cleaves polypeptide chains on the C- terminal side of Met residues, while the trypsin chymotrypsin and enzymes cleave on the C-terminal side of basic (Arg, Lys) and aromatic Phe, Trp, Tyr residues, respectively. With trypsin on digestion, a protein with six Lys and five Arg would yield 12 tryptic peptides, every of that would end with Lys or Arg, apart from the C-terminal peptide. The peptide fragments obtained through specific chemical or enzymatic cleavage are then separated via chromatography example for ion exchange chromatography; and the sequence of each in turn determined through Edman degradation.
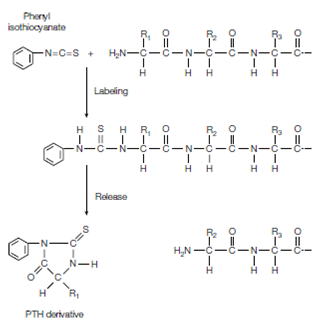
Figure: Edman degradation. The N-terminal amino acid is labeled with phenyl isothiocyanate. Upon mild acid hydrolysis this residue is released as a PTH-derivative and the peptide is shortened by one residue, ready for another round of labeling and release.
While the sequence of every peptide fragment would now be known the order of these fragments in the polypeptide chain would not. The next stage is to produce overlapping fragments through cleaving another sample of the original polypeptide chain with a several enzyme or chemical for instance chymotrypsin then sequencing them and after that separating the fragments. These chymotryptic peptides will overlap one or more of the tryptic peptides enabling the order of the fragments to be build which is shown in figure. In this way the entire length of the polypeptide chain can be sequenced.
To sequence the polypeptides in a multisubunit protein the individual polypeptide chains must first be dissociated through disrupting the noncovalent interactions with denaturing agents like as urea or guanidine hydrochloride. Disulfide bonds in the protein also have to be broken through reduction with 2- dithiothreitol or mercaptoethanol. Iodoacetate is added to form stable S-carboxymethyl derivatives to prevent the cysteine residues recombining. The individual polypeptide chains then have to be separated through, for instance, ion exchange chromatography before sequencing every. Todays, as little as picomole amounts of proteins can be sequenced following their separation through SDS-PAGE either using the polyacrylamide gel containing the protein straightly or following their transfer to nitrocellulose.
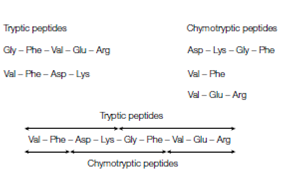
Figure: The use of overlapping fragments to determine the sequence of a peptide. The protein is first digested with trypsin and the resulting peptides separated and sequenced. The protein is separately digested with chymotrypsin and the resulting peptides again separated and sequenced. The order of the peptide fragments in the protein can be determined by comparing the sequences obtained.
To describe the key concepts in the chemical sequencing of a protein are as follows:
1. Purify the protein.
2. Decrease any block and disulfide bonds their re-oxidation with iodoacetate.
3. Cleave the protein with a protease or cyanogen bromide.
4. Separate the fragments and after that sequence them.
5. Look for overlaps among the two collections of sequences in order to construct the full sequence.