RNA splicing:
A key step in RNA processing is the precise removal of intron joining and sequences the ends of neighboring exons to produce a functional mRNA molecule, a procedure known as RNA splicing. The exon-intron boundaries are marked through specific sequences that are shown in the figure. In most cases at the 5' boundary among the exon and the intron (the 5' splice site), the intron begins with the sequence GU and at the 3' exon-intron boundary (the 3' splice site) the intron ends with the sequence AG. A polypyrimidine tract (a conserved stretch of about 11 pyrimidines) lies upstream of the AG at the 3' splice site shows in the figure. Each of these two sequences lies within a longer consensus sequence. A key signal sequence is the branchpoint sequence which is located about 20-50 nt upstream of the 3' splice
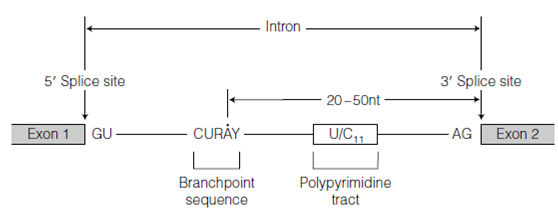
Figure: Conserved sequences for RNA splicing. The residue marked as • in the branchpoint sequence is the site of formation of the 2'5' branch.
site. In vertebrates this sequence is 5' -CURAY-3 where Y pyrimidine and R purine in yeast this sequence is 5' -UACUAAC-3.
RNA splicing occurs in two steps which were described in the above figure. In the first step, the 2' -OH of the A residue at the branch site which is indicated as A in Figure attacks the 3' 5' phosphodiester bond at the 5' splice site causing which bond to break and the 5' end of the intron to loop round and form an commonly 2' 5' bond with the A residue in the branchpoint sequence. Since this A residue already has 3' 5' bonds with its neighbors in the RNA chain the intron becomes branched at this point to form what is called as a lariat intermediate named as like since it resembles a cowboy s lasso. The latest 3'-OH end of exon 1 now attacks the phosphodiester bond at the 3' splice site causing the two exons to release and join the intron, still as a lariat. In each of the two splicing reactions one phosphate-ester bond is exchanged for another for instance: these are two transesterification reactions. Because the number of phosphate-ester bonds is not changed no energy (ATP) is consumed.
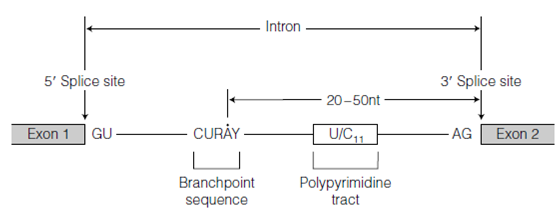
Figure: The two steps of RNA splicing.
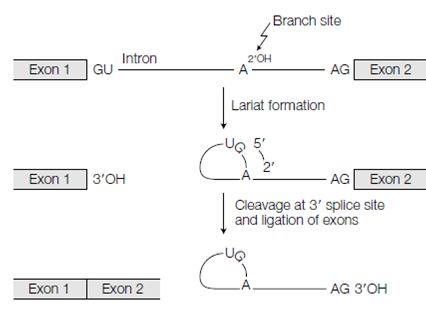
Figure: Formation of the spliceosome.
RNA splicing needs the included of various small nuclear RNAs (snRNAs) every of that is related with various proteins to form a small nuclear ribonucleoprotein particle or snRNP (pronounced 'snurp'). Since snRNAs are rich in U residues they are named as U1, U2, etc. The RNA parts of the snRNPs have regions which are complementary to the 5' and 3' splice site sequences and to other conserved sequences in the intron and so can base pair with them. An U1 snRNP will binds to the 5' splice site and U2 snRNP binds to the branchpoint sequence. A tri-snRNP difficult of U4, U5 and U6 snRNPs then binds as do other accessory proteins, so in which a multicomponent complex that is called a spliceosome is establish at the intron to be erased and causes the intron to be looped out. Therefore, through interactions among the snRNAs and the pre-mRNA the spliceosome brings the downstream and upstream exons together ready for splicing. The spliceosome other catalyzes the two-step splicing reaction to erase ligate and intron together the two exons. The spliceosome then released and dissociates snRNPs can take category in further splicing reactions at other sites on the pre-mRNA.
Although the vast majority of pre-mRNA introns begin with GU at the 5' splice site and end with AG at the 3' splice site, some introns (possibly as many as 1 percent) have variant splice site consensus sequences. In those cases, the intron begins with AU and ends with AC alternate of AU and UG, respectively that is shown in the next diagram. Because RNA splicing includes recognition of the splice site consensus sequences through key
Figure: Comparison of the conserved splice site sequences of the majority of introns (top diagram) with those for AT-AC introns (bottom diagram).
snRNPs, and therefore these sequences are variant in the small intron class, U1, U2, U4 and U6 snRNPs do not take category in splicing these is called AT- AC introns or the AT-AC refers of course to the corresponding DNA sequence. Alternatively, U11, U12, U4atac and U6atac snRNPs are included, exchanges the roles of the U1 and U2, U4 and U6 respectively and assemble to create the AT-AC spliceosome. U5 snRNP is need for splicing both classes of intron.
A few organisms, like as trypanosomes and nematodes, are able to splice both exons from two variant RNA molecules, a procedure called trans-splicing. In this circumstance, the more usual splicing together of two exons in the similar RNA molecule would be cis-splicing.
Some self-splicing introns are also known, for instance, Tetrahymena rRNA and some mitochondrial and chloroplast mRNAs. In that cases the intron RNA sequence catalyzes its own cleavage out of the RNA precursor without the required for a spliceosome. Like catalytic RNA molecules are known as ribozymes a name which is clearly fashioned on 'enzymes', like protein catalysts. The similarity of chemical, some of these self-splicing reactions with the reactions which occur in during spliceosome-mediated splicing has led to a realization of the middle role of RNA catalysis in the latter. The Spliceosome-mediated splicing possibly evolved from self-splicing entities with snRNAs having roles not only in recognition of splice sites but also in the catalytic reactions of spliceosome- mediated splicing. In exacting, the structure formed through U2 snRNA base paired to U6 snRNA probably forms the catalytic middle of the spliceosome.