Cleavage and polyadenylation:
Most of the eukaryotic pre-mRNAs undergo polyadenylation that includes cleavage of the RNA at its 3' end and the although of about 200A residues to form a poly(A) tail. The polyadenylation and cleavage reactions need the existence of a polyadenylation signal sequence that are 5' -AAUAAA-3' located near the 3' end of the pre-mRNA followed through a sequence 5' -YA-3' where Y= a pyrimidine, frequently 5'- CA-3', in the next 11-20 nt that is shown in the figure. The GU-rich sequence or U-rich sequence is also commonly present further downstream. After these sequence parts have been synthesized, two multisubunit proteins known as cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor F (CStF) are transferred from the CTD of RNA polymerase II to the RNA molecule and bind to the sequence components. A protein complex is formed that involves additional cleavage factors and an enzyme known as poly(A) polymerase (PAP). This complex cleaves the RNA among the AAUAAA sequence and the GU-rich sequence which is also described in the given diagram. Poly (A) polymerase then adds about 200A residues to the new 3' end of the RNA molecule using ATP as precursor. As it is build, the poly (A) tail quickly binds multiple copies of a poly (A) binding protein. Poly (A) tail protects the 3' end of the last mRNA against ribonuclease digestion and although stabilizes the mRNA. Additionally, it raises the efficiency of translation of the mRNA. Moreover, some mRNAs, notably histone pre-mRNAs which lack a
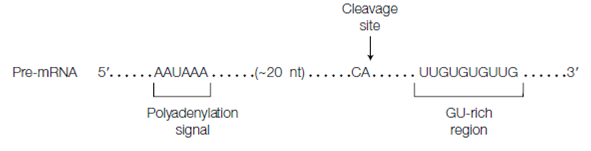
Figure: Conserved sequences for polyadenylation.
Poly (A) tail. Nevertheless, histone pre-mRNA is since subject to 3' processing. It is cleaved near the 3' end through a protein complex which recognizes specific signals, one of that is a stem-loop structure to produce the 3' end of the mature mRNA molecule.