Messenger RNA processing
The common misconception about prokaryotes is that they are unable to perform any processing on their RNA a false distinction often drawn between them and the eukaryotes. Although transcript processing is less common in prokaryotes it still does occur. There is evidence which some bacterial transcripts have short poly-A tails added and there is evidence of intron/exon-like processing in a very few genes. The Archaea possess several of the transcript processing features related with eukaryotes including extensive intron/exon structures but neither the Bacteria nor the Archaea rely heavily on mRNA splicing to generate protein diversity. Alternatively they rely on population diversity to cope with environmental change.
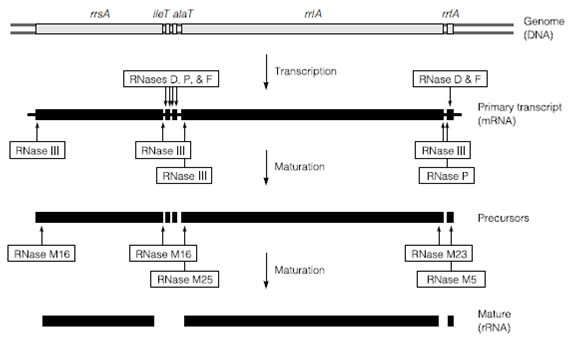
Figure: Processing of the E. coli K12 rrnA primary transcript.
One function in that extensive RNA processing is always present in Bacteria is in the manufacture of the ribosomal RNA. In Escherichia coli there are seven different operons for ribosomal RNA, scattered throughout the genome. The number amount of duplication, location and order of the genes within these operons varies in several species. At the E. coli K12 rrnA genomic locus the 16S rRNA gene is separated from the 23S rRNA gene (rrlA) through genes coding transfer RNA. Downstream of alaT at the end of the operon is the gene for 5S rRNA in Figure. The operon is transcribed as a single polycistronic RNA of about 5000 nucleotides but is quickly processed through RNase III plus other site-specific RNA-hydrolyzing enzymes called M5, M16, M23 and M25. The processing takes place in the order outlined and starts just as the unprocessed RNA emerges from the RNA polymerase example for the 5’ end of the operon is processed before the 3’.