Generalized structure of the bacterial genome
The Bacterial genome is often portrayed as a single, stable, circular molecule. However, the genomes of most Bacteria are fluid constantly changing in response to external stimuli and composed of various molecules including extra chromosomes, megaplasmids and plasmids.
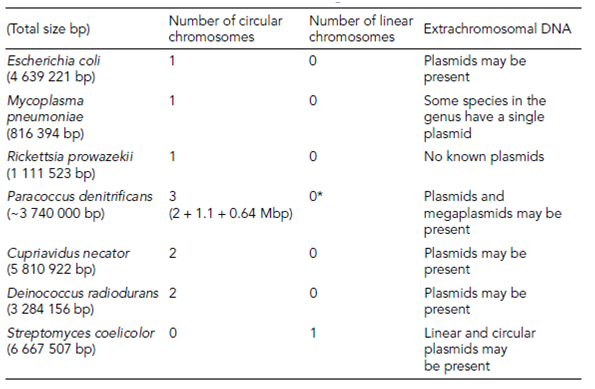
The model organism for molecular biology Escherichia coli is considered to be the paradigm for all Bacterial and Archaeal genomes. However, its single haploid circular chromosome, consisting of around 4.6 million bp, is rather unusual compared with other genera but is through far the best studied. Other Bacterial genomes comprise several chromosomes, some of which are circular and some of which are linear in Table 1.
The size of a Bacterial genome is related to the ecological niche in which the organisms live. Obligate pathogens like as the causative agent of epidemic typhus
Table 1. Chromosomal structures of Bacterial genomes*
seem to have minimized their genomes to such an extent which they rely on host metabolites and proteins in order to replicate. This is taken to the extreme in the smallest known genome in which of Carsonella ruddii that is composed of only 159 663 base pairs of DNA. In comparison, free-living organisms such as the metabolically versatile Pseudomonas aeruginosa and Streptomyces coelicolor have to cope with changes in temperature over tens of degrees varying carbon and energy sources in the space of minutes and other environmental challenges. As a consequence they have a larger complement of genes regulated through a more complex sensing apparatus, and thus a larger genome.
Another strategy used through microorganisms to cope with transient environmental modification is the acquisition of plasmids. Plasmids are small circular extrachromosomal pieces of DNA which replicate independently of the genome. In contrast to the singular genome there may be among 10 and 100 000 complete copies of a plasmid in a Bacterial cell. Plasmids may carry genes which allow the microorganism to become pathogenic, resist antibiotics or metabolize a particular set of compounds. Occasionally these plasmids are integrated into the genome and only exist as extrachromosomal DNA in the presence of certain physiological stimuli. While the plasmids which are used in molecular biology are in the range of 2.5–10 thousand bp (Kbp) naturally occurring plasmids can be various hundreds of thousands of base pairs in size bringing into question the philosophical difference among these megaplasmids and the chromosomes themselves.
The characteristics which distinguish Bacterial genomes from the eukaryotes lie mainly in how the genetic information is arranged. Relatively speaking the Bacterial genome is information-rich containing various regions coding for proteins and RNA but comparatively few regions involved with the regulation of expression. Genes of similar function tend to be clustered together and often genes in a single metabolic pathway or all involved in the synthesis of a complex multi-subunit protein are found in operons. Genes in an operon are sometimes so tightly packed together that they overlap. The fluidity of the Bacterial genome is reflected in gene order found in different Bacterial genera: there is no similarity in the arrangement of genes among the major phyla and often gene order is very different in species of the similar genus.
Different Bacterial genomes have varying composition in terms of nucleotides. The G+C content of the Bacteria ranges from 25 to 75% and this is often reflected in the more frequent use of certain codons for certain amino acids . While Bacterial genomes do contain repeating elements they are often long repeats of >10 bp and may be related with pathogenicity islands insertion sequences or the remnants of excised lysogenic bacteriophage.