Accessory proteins:
DNA polymerases I and III primase and DNA ligase are not the only proteins required for replication of the bacterial chromosome. The DNA sample is a double helix with every strand wound tightly around the other and hence the two strands must be unwound in during replication. How is this unwinding problem resolve? The DNA helicase is used to unwind the double helix (using ATP as energy source) and SSB (single-stranded DNA- binding) protein prevents the single-stranded regions from base pairing again so in which each of the two DNA strands is accessible for replication. In standard, for a replication fork to move along a piece of DNA the DNA helix would require to unwind ahead of it causing the DNA to rotate rapidly. Moreover, the bacterial chromosome is circular and so there are no ends to rotate. The solution to the problem is in which an Enzyme called topoisomerase I breaks a phosphodiester bond in one DNA strand (a single-strand break) a small distance ahead of the fork, permitting the DNA to rotate freely (swivel) around the other intact strand. The phosphodiester bond is then re-formed through the topoisomerase.
When the bacterial circular DNA has been replicated the result is two double- stranded circular DNA molecules which are linked. The Topoisomerase II separates them as follows. This enzyme works in a same manner to topoisomerase I but causes a transient break in every strand (a double-strand break) of a double-stranded DNA molecule. Therefore topoisomerase II binds to one double-stranded DNA causes and circle a transient double-strand break which acts as a 'gate' by that the other DNA circle can pass. The Topoisomerase II then re-seals the strand breaks.
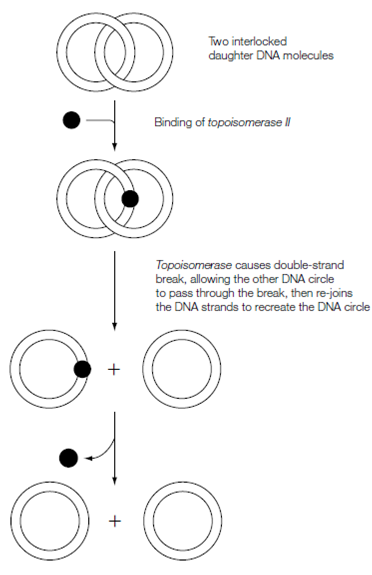
figure: Separation of daughter DNA circles by topoisomerase II.