Cystic fibrosis (CF) is caused by a variety of mutations in the CFTR gene. Imagine you have identified a new single-base pair mutation (A→T) in an exon of the CFTR gene. You wish to use an RFLP approach to determine the proportion of carriers and CF patients (CF patients must have two copies of the defective gene to have the disease) that harbour this particular mutation.
Wild type (normal) CFTR sequence:
5'...
CTCAGATCTGTGATAGAACAGTTTCCTGGGAAGCTTGACTTTGTCCTTGT
GGATGGGGGCTGTGTCCTAAGCCATGGCCACAAGCAGTTGATGTGCTTG
GCTAGATCTGTTCTCAG...3'
(Protein sequence:LRSVIEQFPGKLDFVLVDGGCVLSHGHKQLMCLARSVL)
Mutant CFTR sequence:
5'...
CTCAGATCTGTGATAGAACAGTTTCCTGGGTAGCTTGACTTTGTCCTTGT
GGATGGGGGCTGTGTCCTAAGCCATGGCCACAAGCAGTTGATGTGCTTG
GCTAGATCTGTTCTCAG...3'
a) Mark on the sequences the locations (if present) of
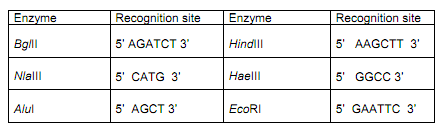
recognition sites for the 6 restriction enzymes listed (BglII, EcoRI, NlaIII, AluI, HaeIII and HindIII)
b) What specific effect will the single base pair mutation have on the CFTR protein?
c) Choose an appropriate pair of enzymes to perform RFLP by southern blot on this sequence; you must select a combination of enzymes that will distinguish the wild type and mutant alleles. Explain your reasoning.
d) Draw the pattern of results you would see following RFLP (using the pair of enzymes chosen above) on a healthy individual, a heterozygous carrier of this mutation, or a cystic fibrosis patient carrying this mutation. NB - assume that the probe used in blotting hybridises to a region from base pair 10 to base pair 105 (i.e. most of the sequence).