Reference no: EM131207511
Evolutionary Biology in the age of Genomics-
Q1. Phylogenetic trees-
You are analyzing sequences sampled from four turtle species (A-D), considering a genomic region that is 75 base pairs long. You observe the following table of the number of substitutions between pairs of species:
|
|
A
|
B
|
C
|
D
|
|
A
|
0
|
9
|
26
|
24
|
|
B
|
|
0
|
25
|
23
|
|
C
|
|
|
0
|
6
|
|
D
|
|
|
|
0
|
A) Construct the phylogenetic tree for these species, assuming the molecular clock.
B) On the basis of the fossil record, you know most recent common ancestor of A & B occurred around 5.4 MYA. Assuming the molecular clock holds, what is your best estimate of the divergence time of C-D? Show your work.
C) New evidence comes to light that suggests that B & C diverged 30 MYA. By assuming the molecular clock for this gene, you estimate that in this time period 50 substitutions should have occurred between B & C. How can you reconcile the expected and observed number of substitutions with the assumption of a constant rate of substitution over the entire phylogeny?
Q2. Population history and trees-
The following diagram shows a history of vicariance events, which have split a population's range into four separate regions over the last 400 million years. You sample DNA from four species from each of the contemporary regions A-D to investigate the relatedness between these populations.
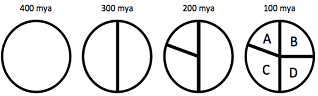
A) Draw a plot with time on the y-axis and indicated the present on top, 100 Mya, 200 Mya, 300 Mya and 400 Mya. Then add a labeled tree for the fours species (A-D), consistent with the lineage splitting being caused by vicariance events, with no subsequent dispersal between them.
B) Analyzing sequence variation from these four species, you tabulate the number of cases in which the pattern is:
A B C D
0 1 1 1
vs
A B C D
1 1 0 1
where 0 is the ancestral allele and 1 the derived allele.
You notice that there is a significant excess of the 1101 configuration. How might you interpret that finding?
C) How would this chronogram look instead if instead species C had arisen from a recent dispersal event from A, i.e., a subset of population A moving to a new location?
D) What type of analyses could you conduct to distinguish between those hypotheses?
Q3. Genetic drift-
You have monitored two populations of snails (A and B) on the west bank of the Hudson river over 100 generations. You genotype these populations at five neutral loci, in every generation, and the allele frequencies in the two populations are plotted below.
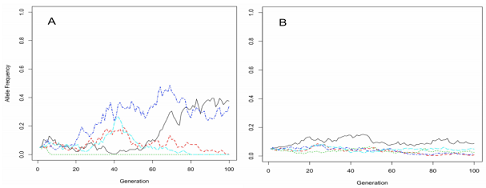
A) Both of these populations are being considered for conservation efforts. A state legislator asks you which of these populations has smaller effective population size, A or B?
B) Which of these populations do you think will have lower levels of genome-wide genetic diversity in the long-term if these trends continue?
C) Briefly justify your answers to parts A and B. Based on your answer, for which species would the conservation effort have a greater chance of success?
D) The blue allele in graph A has an estimated frequency of 30%. What is its expected probability of fixing over the longer term?
Q4. Diversity, divergence-
Human and gorilla diverged 8 million years ago. At a particular gene, which is 1000 bases long, you estimate that substitutions have occurred at 16 sites.
A) Assuming a generation time of 20 years, estimate the substitution rate per site per generation. Show your work.
B) What would you have to assume about mutations that occur within this sequence in order for the substitution rate (per site per generation) to equal the mutation rate (per site per generation)?
C) Sequencing this gene in macaques, you find that the typical individual is heterozygous at 5/1000 sites. What is the effective population size of macaques, assuming that the substitution rate found in part A is indeed the mutation rate? If you did not obtain an answer in A, use a substitution rate of 1x10-9 per site per generation [note that this is not the correct answer to A].
D) This estimated effective population size is a lot lower than the census population size of macaques. What is one plausible explanation of this fact?
5. The Out of Africa model-
A) Why is the observation of low Fst among human populations evidence for the Out of Africa model over the multi-regional theory (in a couple of sentences)?
B) Sequencing a sample of French individuals, you identify that ~1% of the genome consists in regions of approximately 50 kb in which Fst between the French sample and a Nigerian sample is unexpectedly high (~3-fold what is seen elsewhere in the genome). What are two explanations for your findings (1-2 sentences for each)?
C) What additional data could you consider to distinguish between those two hypotheses?
D) Sequencing a remote population on an island of Indonesia, you discover heterozygosity levels (of 0.2% per bp) that exceed what is seen in Africa (typically 0.1-0.12%). In a few sentences, what are the implications for the Out of Africa model?
Q6) Selection-
You are studying the polymorphism that affects flight speed in fireflies. The polymorphism does not appear to affect fecundity.
Homozygotes for the B allele are slow, so only 40% of them survive to have offspring. Heterozygotes for the polymorphism (Bb) fly faster and have a 70% probability of surviving to reproduce. The homozygotes for the other allele (bb) fly very quickly indeed, but often die of exhaustion, with only 10% of them making it to reproduction.
A) What are the relative fitnesses of the three genotypes?
B) The B allele is currently at 95% frequency in the population. Do you expect it to increase or decrease in frequency in the next generation?
C) Calculate the equilibrium frequencies of the two alleles.
D) Calculate the equilibrium frequency of genotypes in adult fireflies. Due to the invasion of owls into the region, flight speed becomes more important for firefly viability. Now 7% of BB flies, 7.7% of Bb flies and 9.1% of bb flies survive to have offspring.
E) What is the probability that the b allele that existed before the owl invasion will fix?
F) If it does, can you calculate approximately how long this will take?
Q7) Individuals who are homozygous for a deletion in the SNAG gene have on average 1.5 children compared to unaffected individuals (all other genotypes), who have on average 2 children. Deletions occur at the gene at the rate of 1x10-5 per generation. At what frequency do you expect to find such deletions in a randomly mating population? Show your work.
Q8) Molecular evolution-
The figure below shows pairwise comparisons of sequences for the gene zamboni between Drosophila melanogaster and three other Drosophila species (A, B and C). The function of this gene is the same in all species. The Y-axis in each panel shows percent conservation of sequence between different species pairs, while the X-axis is position along the gene.
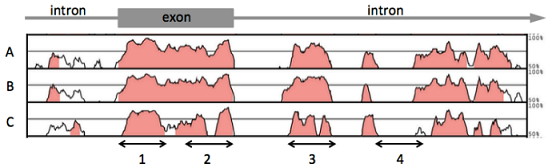
A) Which species would you predict is the most distantly related to D. melanogaster?
B) Which region (1-4) is most likely to be most functionally important?
C) Which non-protein coding region is most likely to harbor a regulatory region for this gene?
Q9) Gene duplications-
Species 3 is an outgroup to Species 1 and 2. Consider three duplicate genes, A, B, and C in Species 1, 2, and 3. All three species have gene A. Gene B arose as a duplicate of gene A, and now only occurs in Species 1 and 2. Gene C is a duplicate of B and occurs only in Species 2.
A) Assume no gene losses. Mark the location of each duplication on the species phylogeny below, indicating which gene was duplicated:
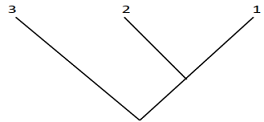
B) Now draw and label the gene tree consistent with the above information. Denote each tip with the letter of the gene followed by the number of the species (ie, A1, A2, A3, B1, B2, & C2)
You study the expression of all of the genes in the three species and mark the expression of the gene with an X:
|
|
Hind-Limb
|
Force-Limb
|
Brain
|
|
A1
|
|
X
|
|
|
B1
|
X
|
|
|
|
A2
|
X
|
X
|
|
|
B2
|
X
|
X
|
X
|
|
C2
|
X
|
X
|
|
|
A3
|
X
|
X
|
|
C) Fill in the blanks:
Gene________ in species_________ is best described as having undergone neofunctionalization.
Genes________ and ________ in species_________ are best described as having undergone subfunctionalization.
D) Regulation of these genes involves a trio of transcription factors.
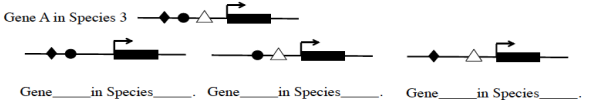
The circle transcription factor is expressed in the brain and the hind-limbs, the diamond transcription factor is expressed in the brain and fore-limbs. Both are activators of gene expression. The triangle transcription factor is expressed only in the brain and is a repressor (if the regulatory region is bound by this factor it will not be expressed). Below we show the regulatory region of some of these genes, showing the binding sites for the three different transcription factors. The regulatory region of the gene A in Species 3 is labeled. Identify the three other genes shown below:
Q10) Speciation-
A) A researcher wishes to use the biological species concept to categorize a clade of newly discovered, asexual desert lizards. Do you agree with their decision? Why?
B) Under allopatric models of speciation is reproductive isolation an active or passive consequence of selection? Briefly explain your answer.
C) Do differences in ecology between a pair of populations increase or decrease the rate of accumulation of reproductive isolating factors under an allopatric speciation model? Briefly explain your answer.
11) Speciation-
You are studying two species pairs of oaks that are now broadly sympatric. Species pair A are reproductively isolated from each other, as the hybrids produce acorns at the wrong time of year and are also partially sterile. Species pair B are reproductively isolated from each other, as they flower at different times of year, and the hybrids are more susceptible to disease.
A) In which species pair is reinforcement more likely to have played a role in speciation? Briefly explain your answer.
B) Which barrier may have evolved through reinforcement? Briefly explain your answer.
C) What does your reinforcement hypothesis predict about this trait for individuals sampled where the species pair is allopatric?
D) What are Coyne & Orr's (1989) main results that suggest that support the occurrence of reinforcement rather than Templeton's alternative explanation? Briefly explain.
Q12) Speciation-
You work at the NY state avian conservation program and are interested in a clade of hummingbirds. Members of this clade of hummingbirds have 14 chromosomes, where chromosome 12 is a heteromorphic sex chromosome. Males have two normal copies of chromosome 12, while individuals who carry one degenerate copy of chromosome 12 are females.
A) You colleague tells you that one sex of hybrid between two species is sterile due to failures in gametogenesis, while the other sex is fertile. Which sex do you think is sterile?
B) You are a reviewer for a paper submitted to PLOS Biology which claims that sex-specific hybrid sterility between these two species is caused by an incompatibility between alleles at two loci on chromosomes 3 and 8. Do you believe this claim? Please explain your answer.