Reference no: EM131126738
1. The majority of FDA approved drugs can be profitably classified into three primary groups:
a. small molecules, peptides and antibodies
b. lipids, carbohydrates and nucleic acids
c. peptides, nucleic acids and lipids
d. organic, inorganic and biological
2. Aileron Therapeutics is developing ____________ to treat various diseases.
a. non-peptidic macrocycles
b. antibodies
c. stapled peptides
d. Unconstrained peptides
3. Identify the three companies with in silico platform technologies that are also correctly matched with drug modality (in parentheses):
a. Ironwood (peptides), Aileron (peptides) and Ensemble (macocycles)
b. Zymeworks (peptides), Nimbus Discovery (peptides) and Pieris (small molecules)
c. CytomX (antibodies), Circle Pharma (peptides) and Numerate (small molecules)
d. Numerate (small molecules), Circle Pharma (macrocyclic peptides) and Zymeworks (antibodies)
4. Some proposed advantages of peptide-based drugs compared to small molecule and antibody drugs, respectively, include:
a. improved toxicity profile and potential for cell penetration
b. targeting of protein-protein interactions and larger size
c. targeting of enzyme-substrate interactions and larger size
d. oral bioavailability and proven track record
5. Peptides drug candidates can often be identified using a bacteriophage-based library screening method called ______ . In a typical screening experiment greater than _______ peptides can be screened for binding or activity against a protein target, resulting in confirmed hit rates of about ________. The company ______ employs ______ as its primary drug discovery technology.
a. yeast two hybrid, 100, 70%, Lysogene, yeast two hybrid
b. chemical HTS, 1000, 35%, Ra Pharmaceuticals, HTS
c. phage display, 109, much less than 1.0%, Dyax, phage display
d. phage display, 109, much greater than 50.0%, Achillion , phage display
6. The drug development process can be divided into four consecutive stages:
a. clinical testing, target validation, sales and marketing and regulatory approval
b. target validation, discovery, pre-clinical development and clinical testing
c. chemistry, physics, biology and financial analysis
d. discovery, pre-clinical development, clinical testing and target validation
7. A primary purpose of randomization in a RCT is to:
a. establish inclusion criteria
b. establish exclusion criteria
c. enable the use of calculus to analyze trial results
d. eliminate selection bias and confounding
8. The primary uses of computational tools, like YASARA, in drug discovery include:
a. visualization and design
b. visualization, analysis, modeling, design and simulation
c. researching articles
d. preparing presentations
9. Three freely available (for academic use) modeling programs that can be used for drug design include:
a. Deep View, ArgusLab, and Sybyl
b. Deep View, Glide, and MOE
c. Discovery studio, YASARA, and MOE
d. SPDBV, VegaZZ, and ArgusLab
10. Three companies that specifically sell drug design software include:
a. Microsoft, Apple and Linux
b. Tripos, Schrodinger and OpenEye
c. Aspen Avionics, Fuji and Unix
d. Linux, Unix and Bio-Rad
11. the four main classes of biomolecules are:
a. co-factors, ions, vitamins and minerals
b. lipids, proteins, nucleic acids and water
c. polypeptides, nucleic acids, polysaccharides and lipids
d. amino acids, nucleotides, monosaccharides and fatty acids
12. ____ are the monomeric building blocks of proteins and peptides. There are _____ common or standard ________ with ___ chirality. They are often classified according to the nature and properties of their ______.
a. nucleotides, 25, hydrogen bonds, Z, bases
b. nucleotides, 25, phosphate groups, X, bases
c. amino acids, 20, amino acids, L, main chains
d. amino acids, 20, amino acids, L, side chains
e. amino acids, 20, amino acids, D, side chains
13. Which amino acid do you think will migrate most towards the cathode (negative side) when separated by electrophoresis at pH 7.1?
a. E
b. Tyr
c. Ala
d. K
14. Calculate the approximate pH at which Lysine will tend not to move in an applied electrical field.
a. 3.2
b. 14.6
c. 5.7
d. 9.9
15. Nucleic acids (DNA and RNA) are polymers of ______.
a. carbohydrates
b. fatty acids
c. nucleotides
d. nucleosides
16. The ____are the primary sites of cellular energy metabolism which includes three primary catabolic pathways referred to as ____, ____, and ____. The main chemical end products of ceullar respiration include ____, ____ and ____.
a. ribosomes, glycolysis, gluconeogenesis, citirc acid cycle, water, AMP and oxygen
b. ribosomes, glycolysis, krebs cycle, electron transport chain, water, ATP and carbon dioxide
c. mitochondria, glycolysis, krebs cycle, electron transport chain, water, ATP and carbon dioxide
d. mitochondria, glycolysis, krebs cycle, electron transport chain, water, ATP and carbon monoxide
17. Assuming physiological conditions, the peptide ARLSKQ would tend to be more soluble than each of the following peptides (True (T) or False (F)).
a. ALVFIA, assuming a pH of 6.5
b. ARLSKQ, assuming a pH of 12.0
c. VRLIKF, assuming a pH of 7.0
18. Assume you have the following monosaccharide reactants, all in cyclized form: L-glucose, L-mannose, L-galactose, L-fructose and L-ribulose. Further assume that they can only be combined into linear tetra-saccharide polymers with alpha-glycosidic linkages. Given these assumptions, how many mathematically possible tetra-saccharides are there?
a. 1024
b. 625
c. 40
d. 3125
e. None of the above
19. Proteins can move laterally in biomembranes and the lateral distance (r) traveled in a given time (t) is given by,r=(4Dt)1/2. The diffusion constant for (D) for the membrane protein fibronectin is approximately 0.7 x 10-12 cm2•sec-1, whereas that for rhodopsin is about 3.0 x 10-9 cm2•sec-1.
a. calculate the distance (r) traveled by each of these proteins in 10 msec (answer in nm; be sure to circle your answers)
b. what can you reasonably assume about the interactions of these two proteins with other membrane components? Your answer should be 1-4 sentences.
20. Two drug candidates were tested in a cell-based assay. Addition of drug candidate 1 resulted in an increase in intracellular cAMP. Addition of drug candidate 2 also resulted in an increase in cAMP Chemical and structural analysis indicated that both drug candidates have a doubly hydroxylated benzene ring that is connected to a linear -C(OHH)-C(H2)-NH-CH3 tail. Interestingly, drug candidate 1 produced even more dramatic increases in cAMP levels when co-administered with a cyclic nucleotide phosphodiesterase inhibitor. This effect was not observed for drug candidate 2. Addition of drug candidate 1 also increased the phosphorylation status of key cellular signaling proteins. This effect was not observed for drug candidate 2. Based on this data and material presented in lecture, what receptor do you think drug candidate 1 is acting on? Do you think drug candidate 2 is acting on the same receptor. Assuming satisfaction of the Lipinski rules reliably predicts oral bio-availability, would you predict drug candidate 1 to be orally active? Please support your answers with facts and logical arguments.
21. You are tasked with identifying lead molecules for a protein tyrosine kinase called PTKA1. All lead molecules must be highly specific for inhibition of PTKA1 over its isoform PTKA2. More specifically, any lead molecule must have at least a low nano-molar IC50 against PTKA1 and be 10x more potent an inhibitor of PTKA1 than PTKA2. After running a PTKA1 screen and PTKA2 counter-screen, you obtain the following results. Circle all potential lead molecules.

22. Two proteins are being proposed as novel metabolic cancer targets. One target is a lactic acid transporter called MCT1. The other target is an enzyme complex called the pyruvate dehydrogenase (PDH) complex. The PDH complex catalyzes the conversion of pyruvate to acetyl-CoA:
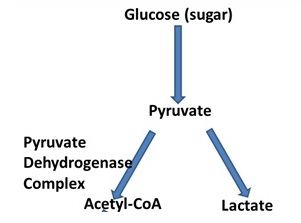
The Results from biological testing of cancerous tumors suggest that under aerobic and anaerobic conditions, extracellular lactate levels correlate with cancer aggressiveness and resistance to chemotherapy. Clinical results further indicate differential up-regulation of MCT1 and down-regulation of MCT4 (another lactate transporter) on aggressive and treatment-resistant tumors. Preliminary pre-clinical results from healthy mice suggest MCT4 knockouts (but not MCT1 knockouts) are lethal. Based on the two proposed targets and the evidence provided, what type of drug should be developed? Please circle the correct answer.
a.PDH inhibitor
b. MCT1 and MCT4 inhibitors
c. MCT1 inhibitor
d. MCT4 inhibitor
23. Consider graphs A, B, C, and D. All four graphs involve the same protein receptor. Graph A and D involve the exact same receptor substrate (ligand). Graph B involves one drug (drug), while graphs C and D involve a second drug (drug 2). Graph D involves drug 2 being tested at different doses.
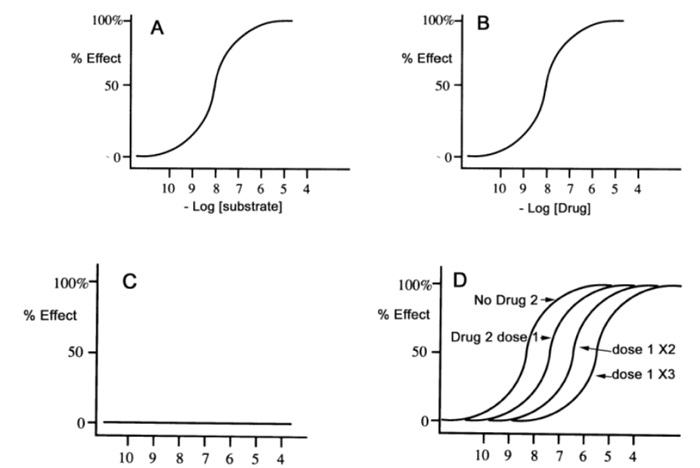
With reference to all the graphs, circle the correct answer.
a. The drug in graph B is an agonist that is just as potent as the natural ligand. Drug 2 is an antagonist of the same receptor.
b. The drug in graph B is an agonist that is less potent than the natural ligand. Drug 2 is an antagonist of the same receptor.
c. The drug in graph B is an agonist that is more potent than the natural ligand. Drug 2 is an antagonist of the same receptor.
d. The drug in graph B is an agonist that is just as potent as the natural ligand. Drug 2 is an agonist of the same receptor.
e. The drug in graph A is an agonist that is just as potent as the natural ligand in graph B. Drug 2 is an antagonist of the same receptor.
f. None of the above
24. Consider the following SAR dataset in the table below. All the data was collected at pH = 7.0 unless otherwise stated.
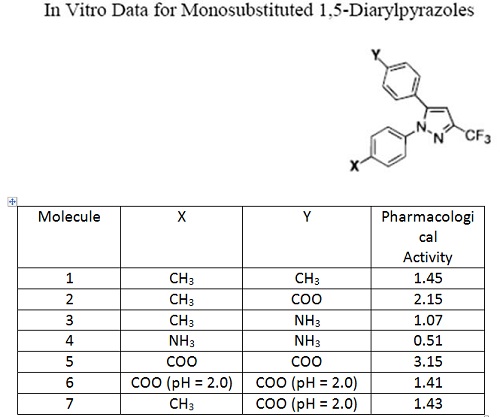
A reasonable inference from the data is that,
a. pharmacological activity increases with molecular positive charge
b. pharmacological activity decreases with molecular negative charge
c. pharmacological activity increases with molecular volume
d. pharmacological activity increases with molecular negative charge
e. none of the above
25. LIGPLOT is a program for visualizing and analyzing non-covalent protein-ligand interactions. Consider the following LIGPLOT schematic for the ligand ZINCCO9831589 and its predicted interaction with the active site residues of the enzyme O-acetyl-L-serine sulfhydrylase.
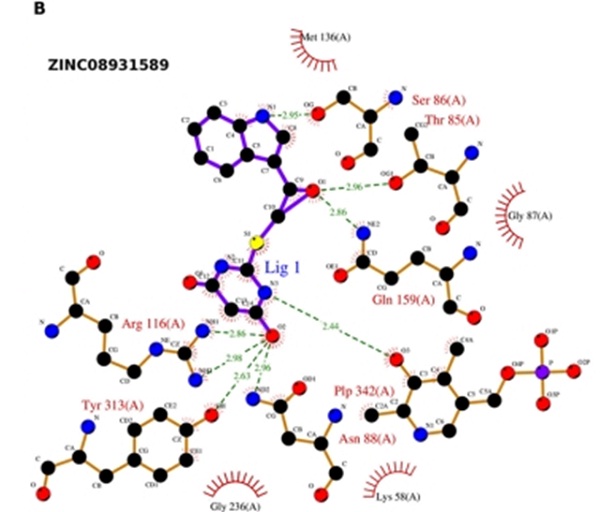
Assume enzyme-ligand hydrogen bonds and buried hydrophobic ligand surface area (500 Å2) determine the free energy of binding between the enzyme and ligand. Furthermore, assume each hydrogen bond contributes -0.9 kcal/mol to the binding free energy and that the buried ligand hydrophobic surface area contributes -0.005 kcal/ Å2 mol to the binding free energy. Assume RT ≈ 0.6 kcal/mol. Based on the provided data, calculate the predicted binding free energy and dissociation constant for enzyme-ligand binding. Please circle the correct answer.
a. -15.5 kcal/mol, 8.51 x 105
b. -9.5 kcal/mol, 1.54 x 105
c. -9.5 kcal/mol, 9.53 x 10-8
d. -3.5 kcal/mol, 9.73 x 10-8
e. +12.5 kcal/mol, 3.12 x 10-12